Fengming Lin
Postdoctoral Research Associate | Computational Medicine & Generative AI | School of Computer Science | The University of Manchester

Christabel Pankhurst Institute, Oxford Road, University of Manchester, M13 9PL Manchester
I am Fengming Lin (林枫茗), a Postdoctoral Research Associate at the University of Manchester, working in the field of Computational Medicine and Generative Artificial Intelligence.
I received my PhD in Computer Science from the University of Leeds, supervised by Prof. Alejandro F. Frangi, Prof Yan Xia, and Dr Nishant Ravikumar. My doctoral research focused on artificial intelligence for vascular segmentation and modality-generalized aneurysm detection. Prior to that, I obtained my Master’s and Bachelor’s degrees from Shandong University, where I was supervised by Prof. Ju Liu and Prof. Qiang Wu, focusing on deep learning for medical image analysis.
My research interests lie at the intersection of AI, medical image analysis, and computational modeling, with applications to:
- Image segmentation, registration, and generative modeling
- Digital twin and virtual population construction
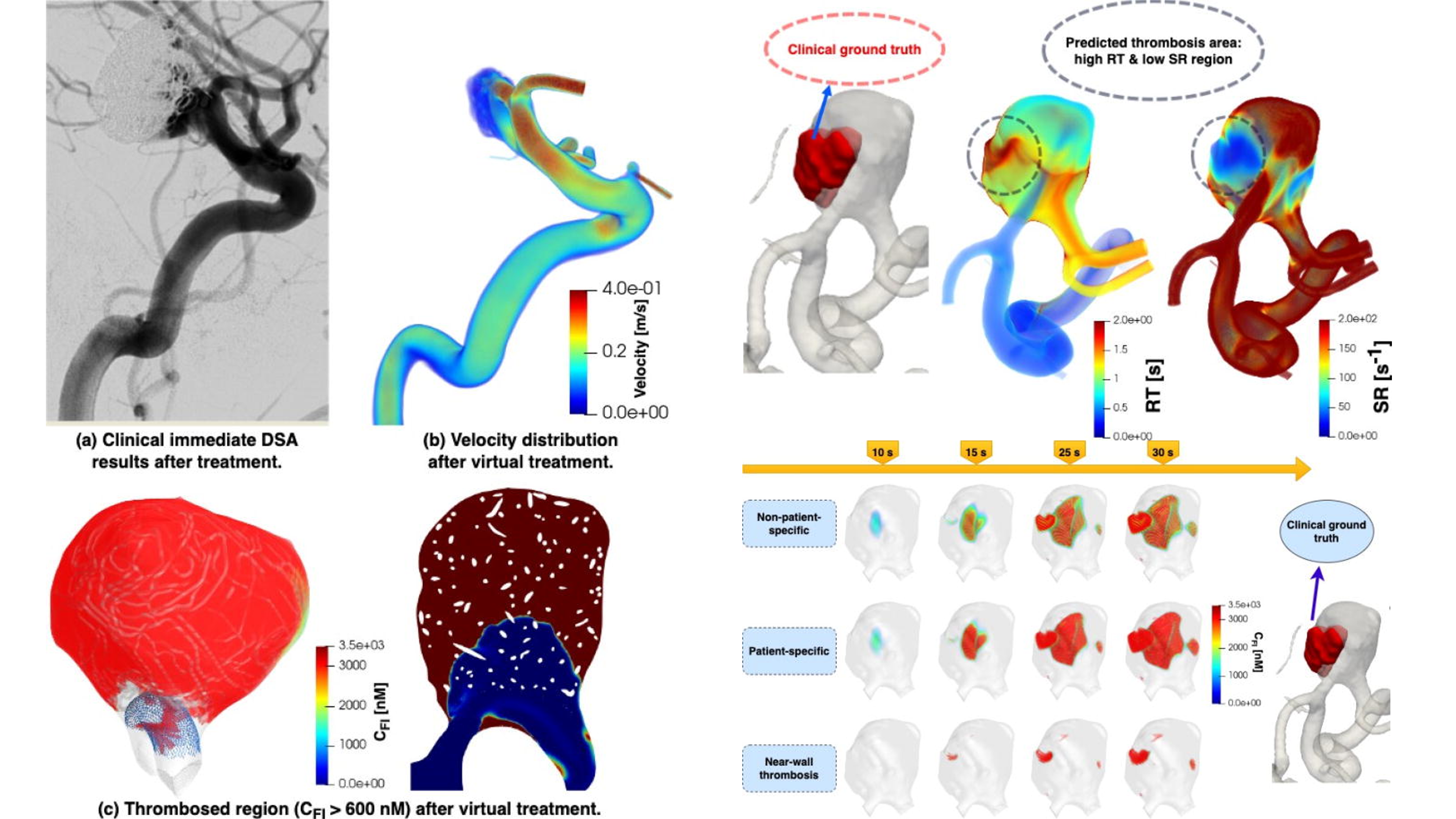
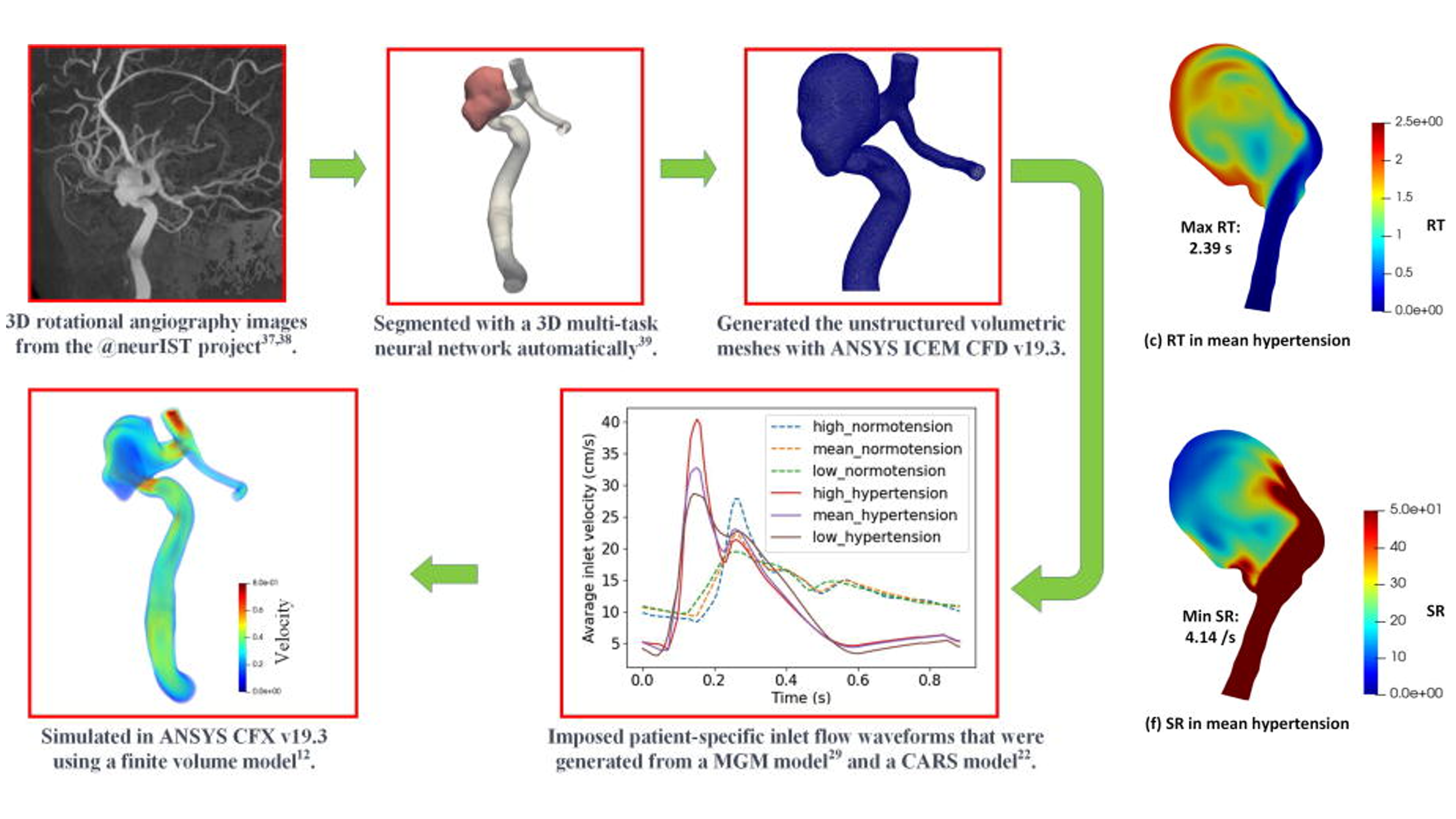
- In-silico trials for cerebral vascular and cardiovascular devices
- Multi-modal and domain-generalizable deep learning methods
I published a few papers in journals and conferences that you might not have heard of such as Computer Methods and Programs in Biomedicine (CMPB) and IEEE ISBI on Google Scholar.
I am actively involved in international collaborations on fluid dynamics modeling, generative models, and large language models for medical applications. Beyond research, I also serve as a reviewer for journals such as Medical Image Analysis, IEEE TMI, IEEE TNNLS, IEEE TAI, and PR.
My long-term goal is to advance the development of Fully automated in-silico clinical trials, bridging computational science and healthcare innovation.
news
| Jun 16, 2025 | Website launch announcement: the new website is now live! |
|---|
selected publications
-
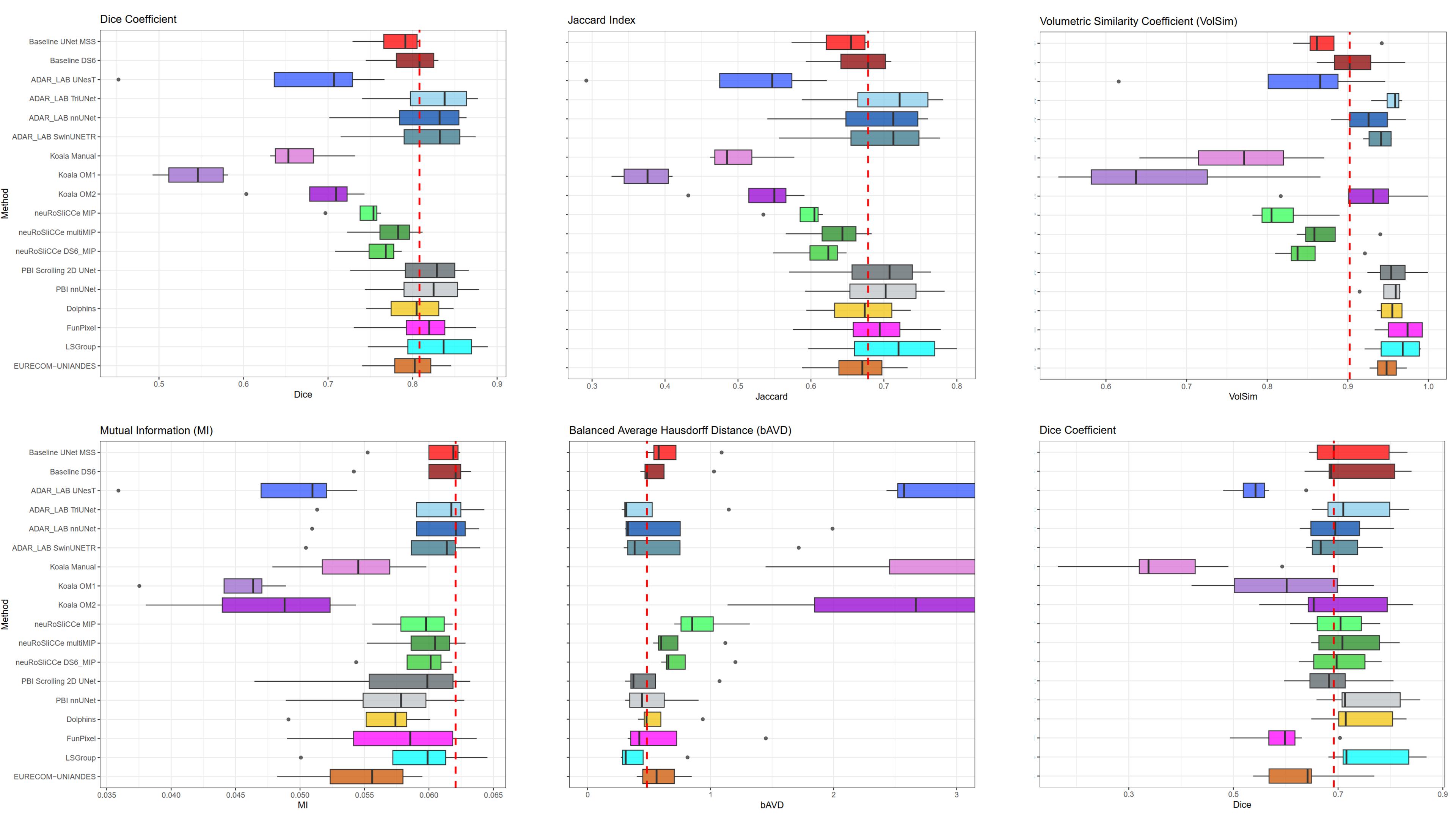
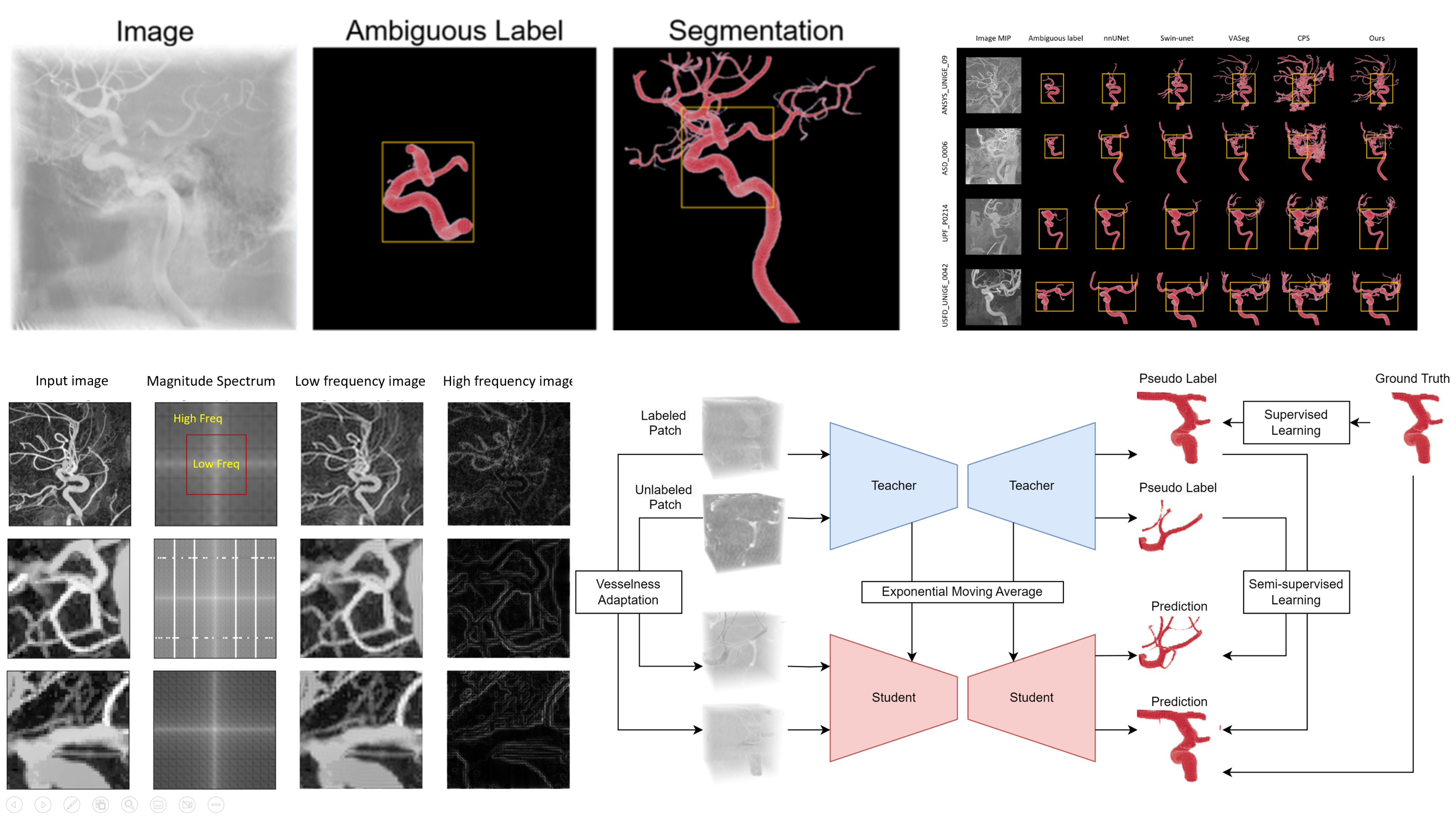 (2023). Adaptive semi-supervised segmentation of brain vessels with ambiguous labels. International Conference on Medical Image Computing and Computer-Assisted Intervention:106–116.
(2023). Adaptive semi-supervised segmentation of brain vessels with ambiguous labels. International Conference on Medical Image Computing and Computer-Assisted Intervention:106–116. -
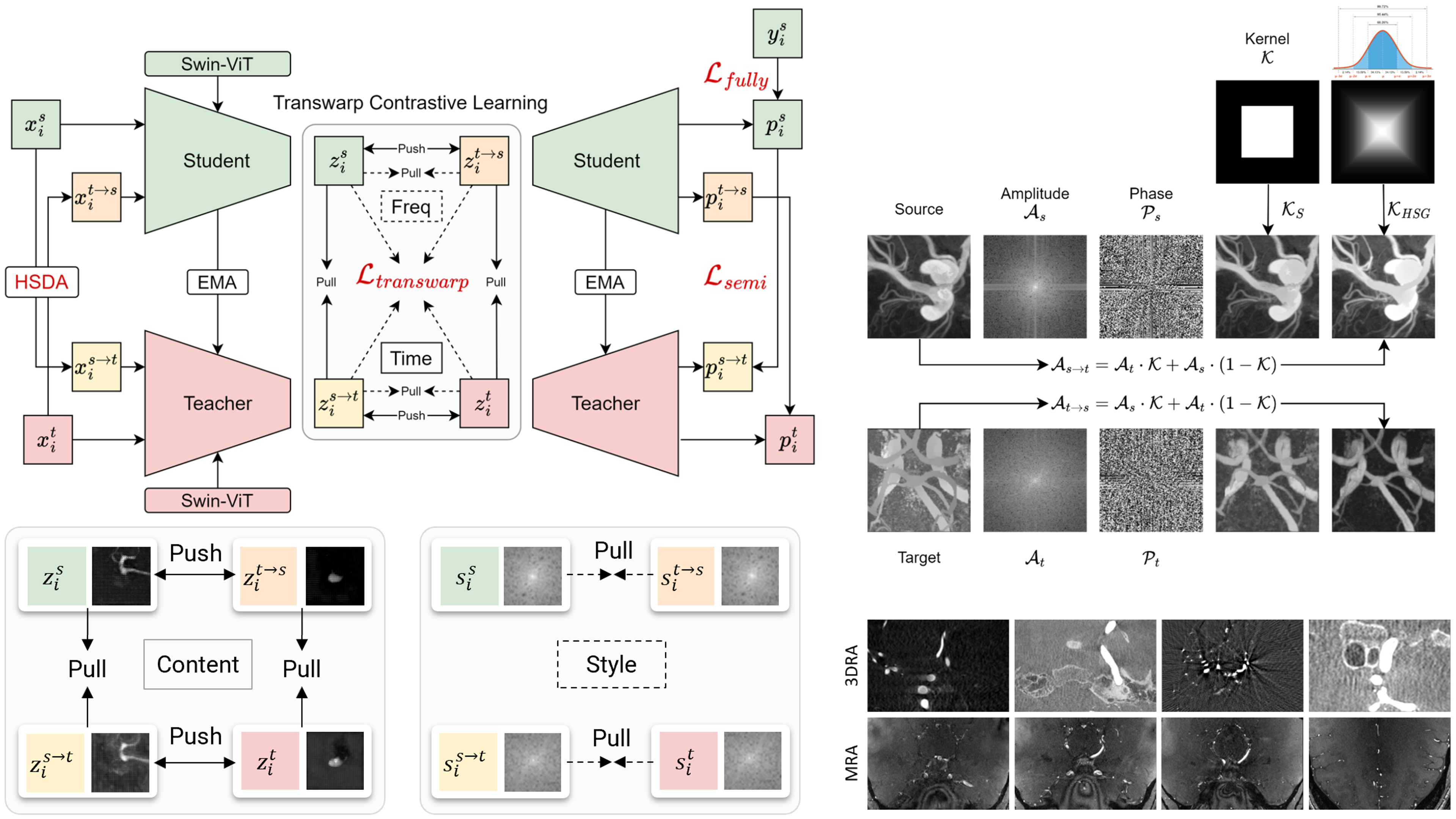 (2024). Unsupervised Domain Adaptation for Brain Vessel Segmentation Through Transwarp Contrastive Learning. 2024 IEEE International Symposium on Biomedical Imaging (ISBI).
(2024). Unsupervised Domain Adaptation for Brain Vessel Segmentation Through Transwarp Contrastive Learning. 2024 IEEE International Symposium on Biomedical Imaging (ISBI). -
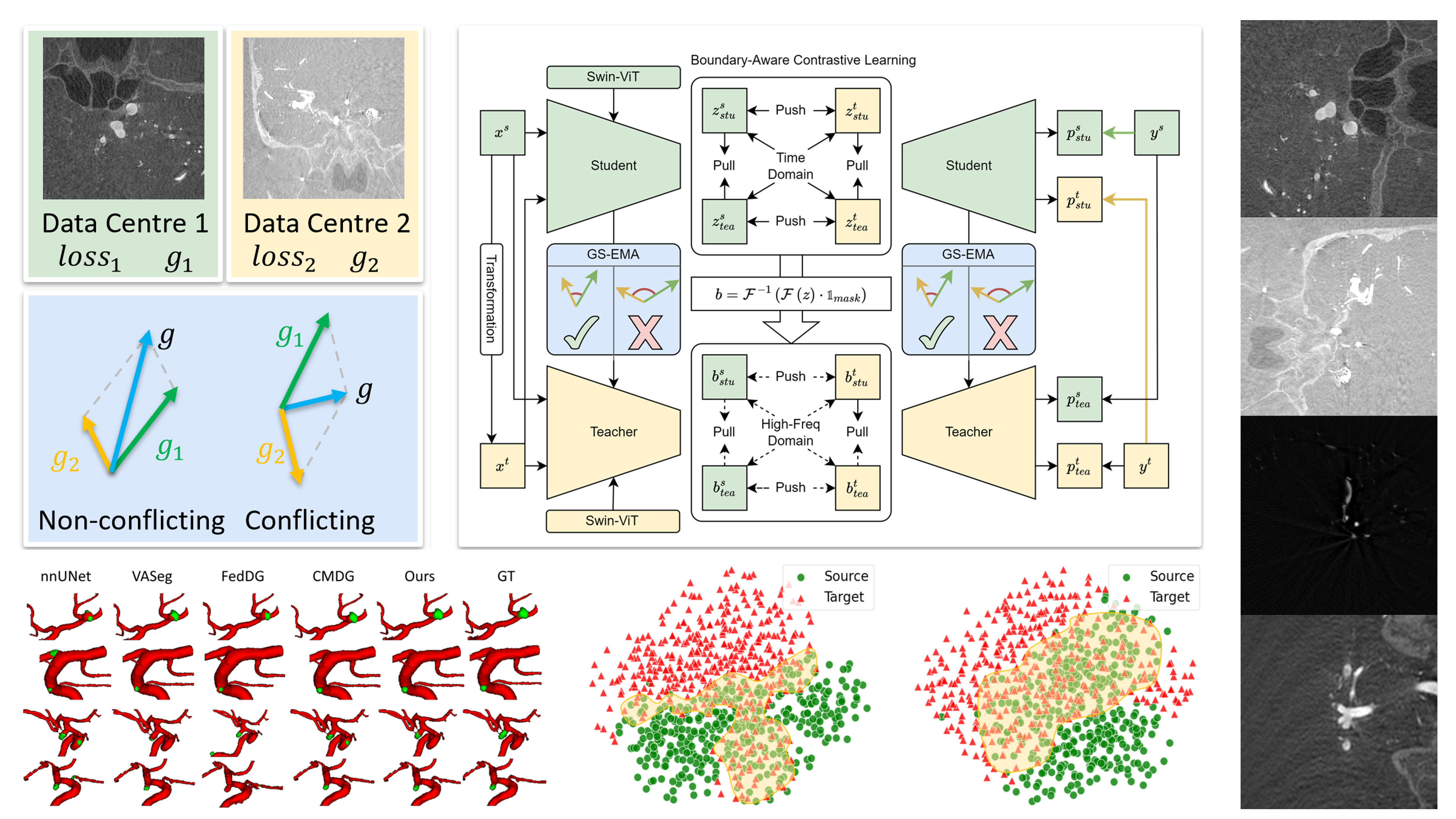 (2024). GS-EMA: Integrating Gradient Surgery Exponential Moving Average with Boundary-Aware Contrastive Learning for Enhanced Domain Generalization in Aneurysm Segmentation. 2024 IEEE International Symposium on Biomedical Imaging (ISBI).
(2024). GS-EMA: Integrating Gradient Surgery Exponential Moving Average with Boundary-Aware Contrastive Learning for Enhanced Domain Generalization in Aneurysm Segmentation. 2024 IEEE International Symposium on Biomedical Imaging (ISBI). -
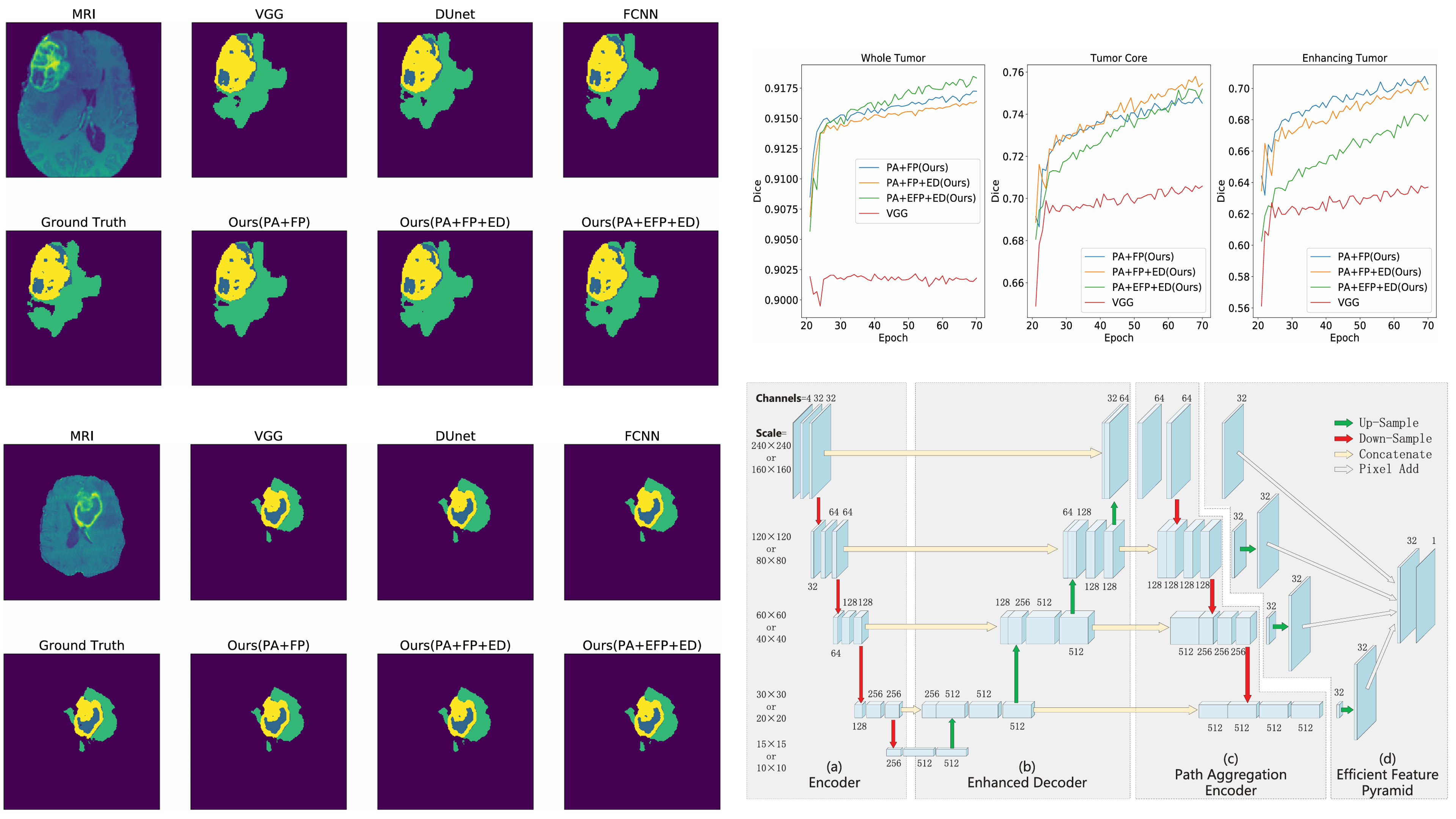
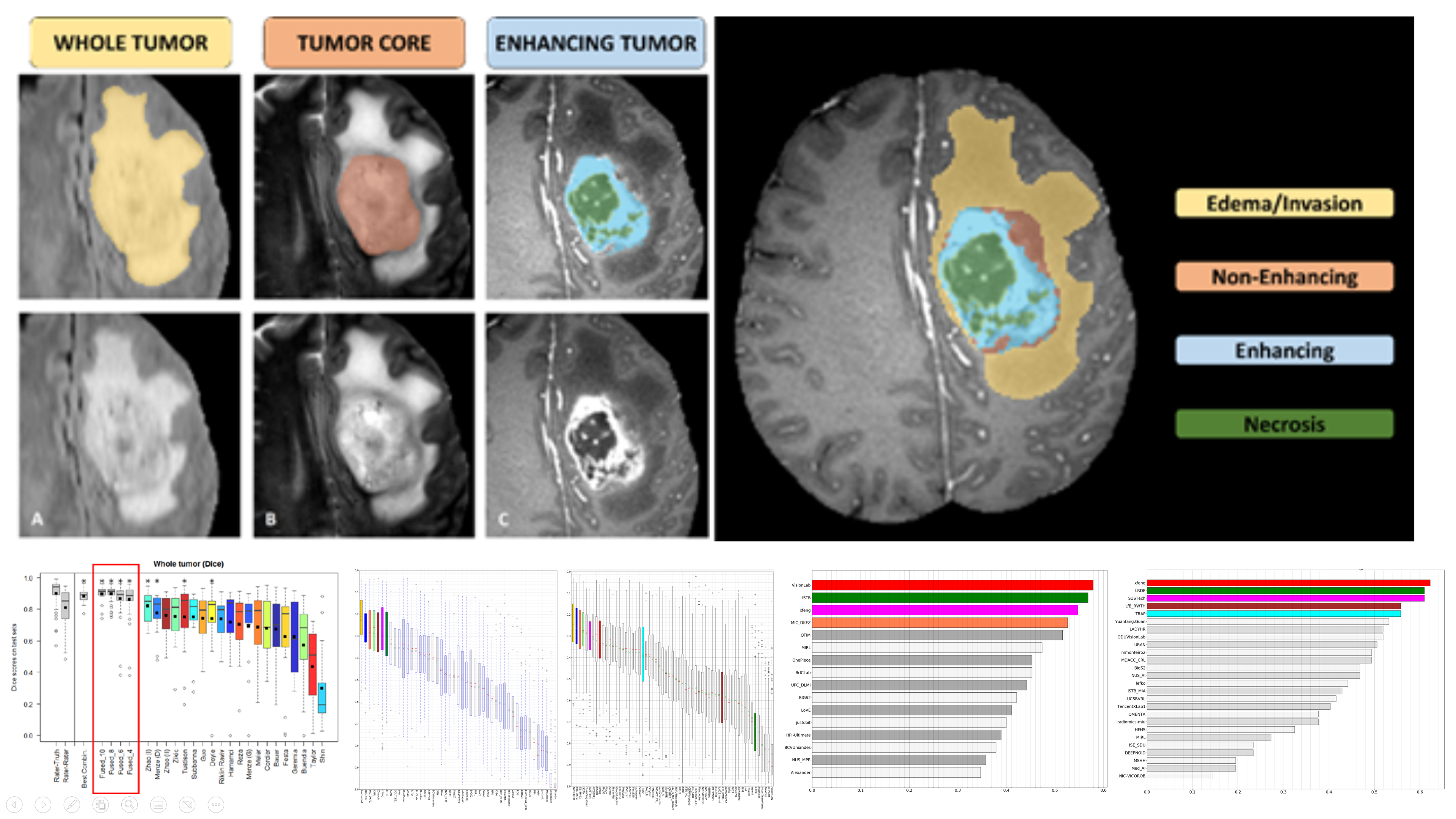
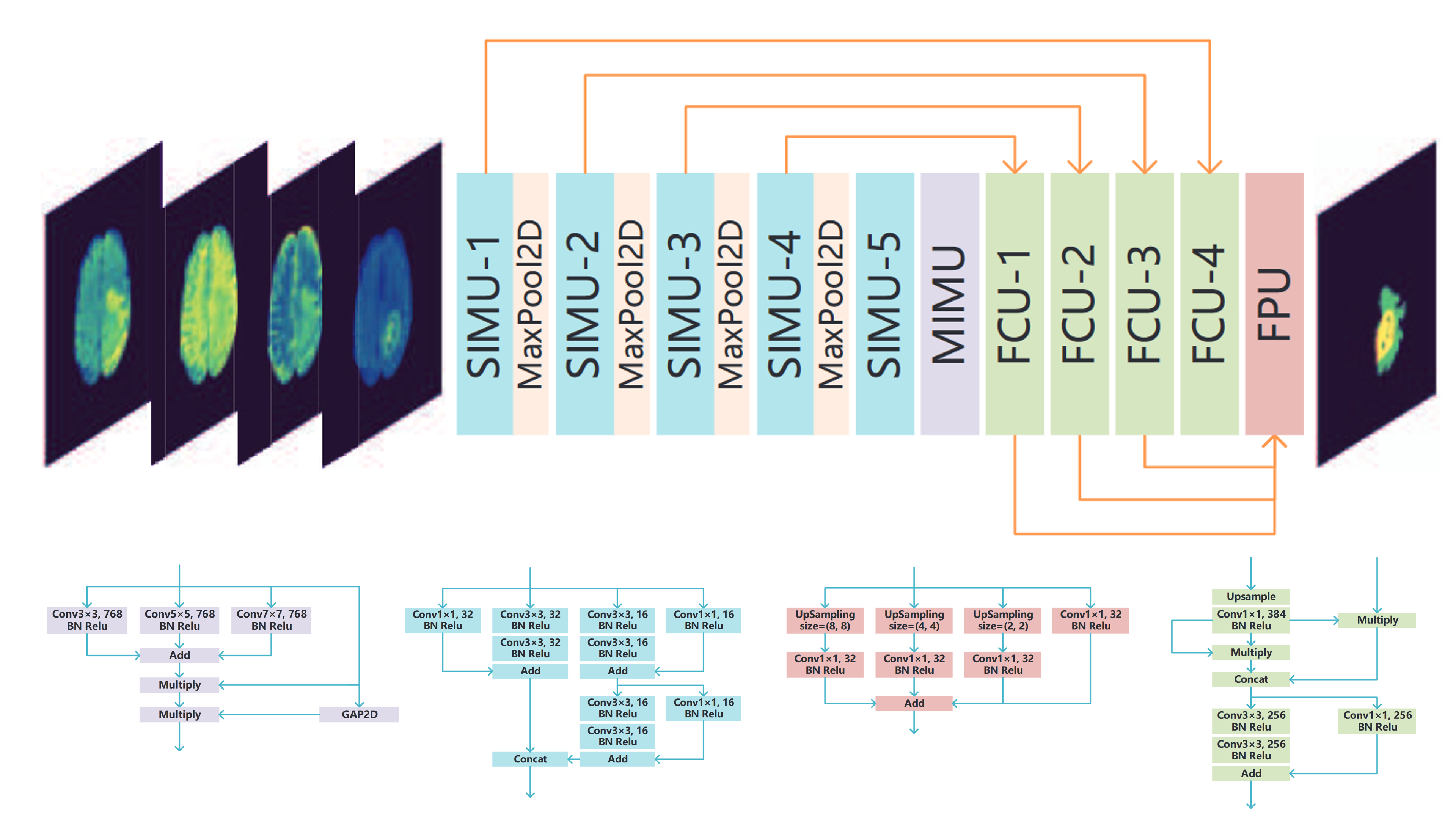 (2019). FMNet: feature mining networks for brain tumor segmentation. 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI):555–560.
(2019). FMNet: feature mining networks for brain tumor segmentation. 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI):555–560. -
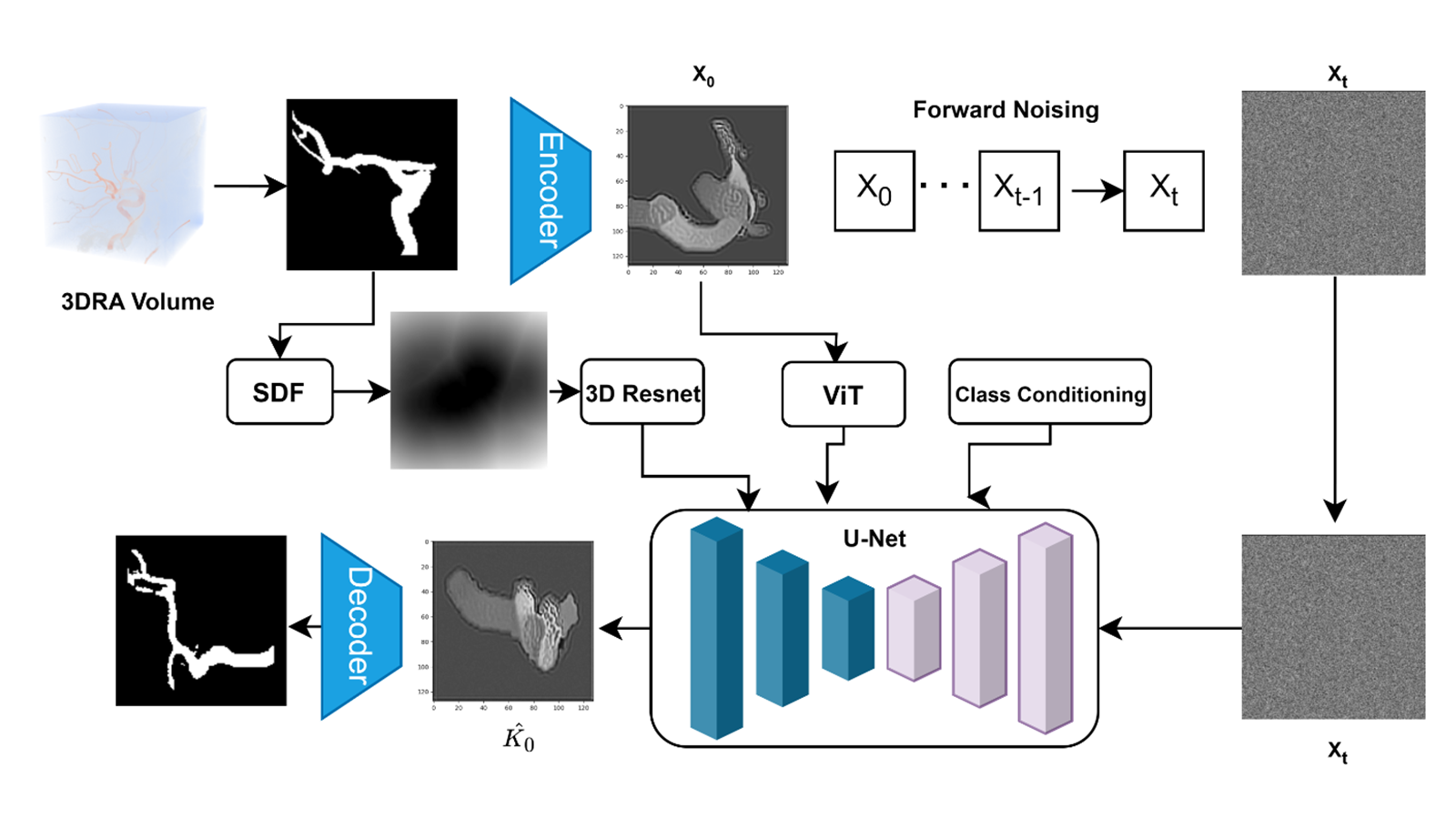 (2024). Few-shot learning in diffusion models for generating cerebral aneurysm geometries. 2024 IEEE International Symposium on Biomedical Imaging (ISBI).
(2024). Few-shot learning in diffusion models for generating cerebral aneurysm geometries. 2024 IEEE International Symposium on Biomedical Imaging (ISBI). -
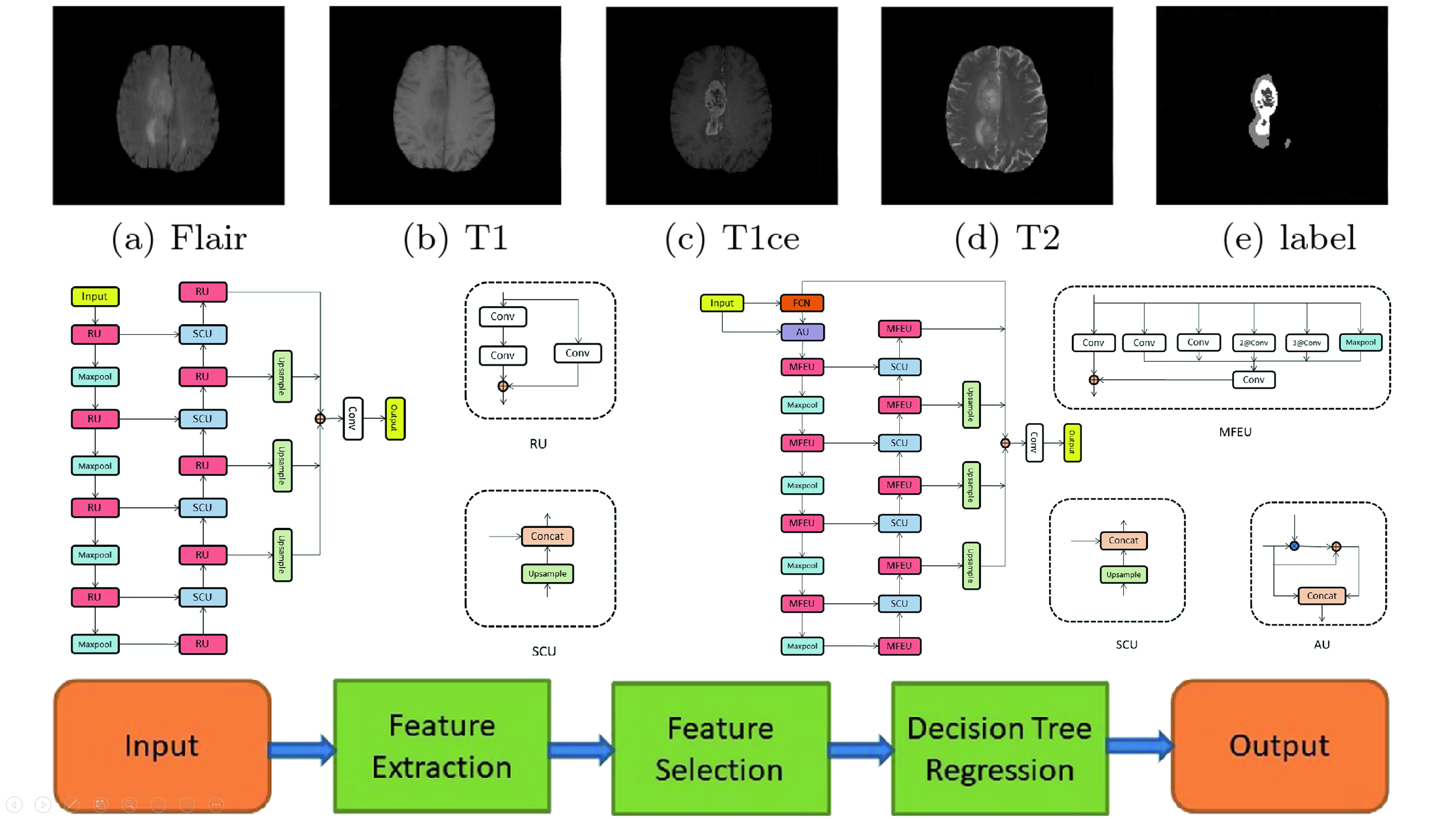 (2019). Brain tumor segmentation using dense channels 2D U-Net and multiple feature extraction network. International MICCAI brainlesion workshop:273–283.
(2019). Brain tumor segmentation using dense channels 2D U-Net and multiple feature extraction network. International MICCAI brainlesion workshop:273–283. -
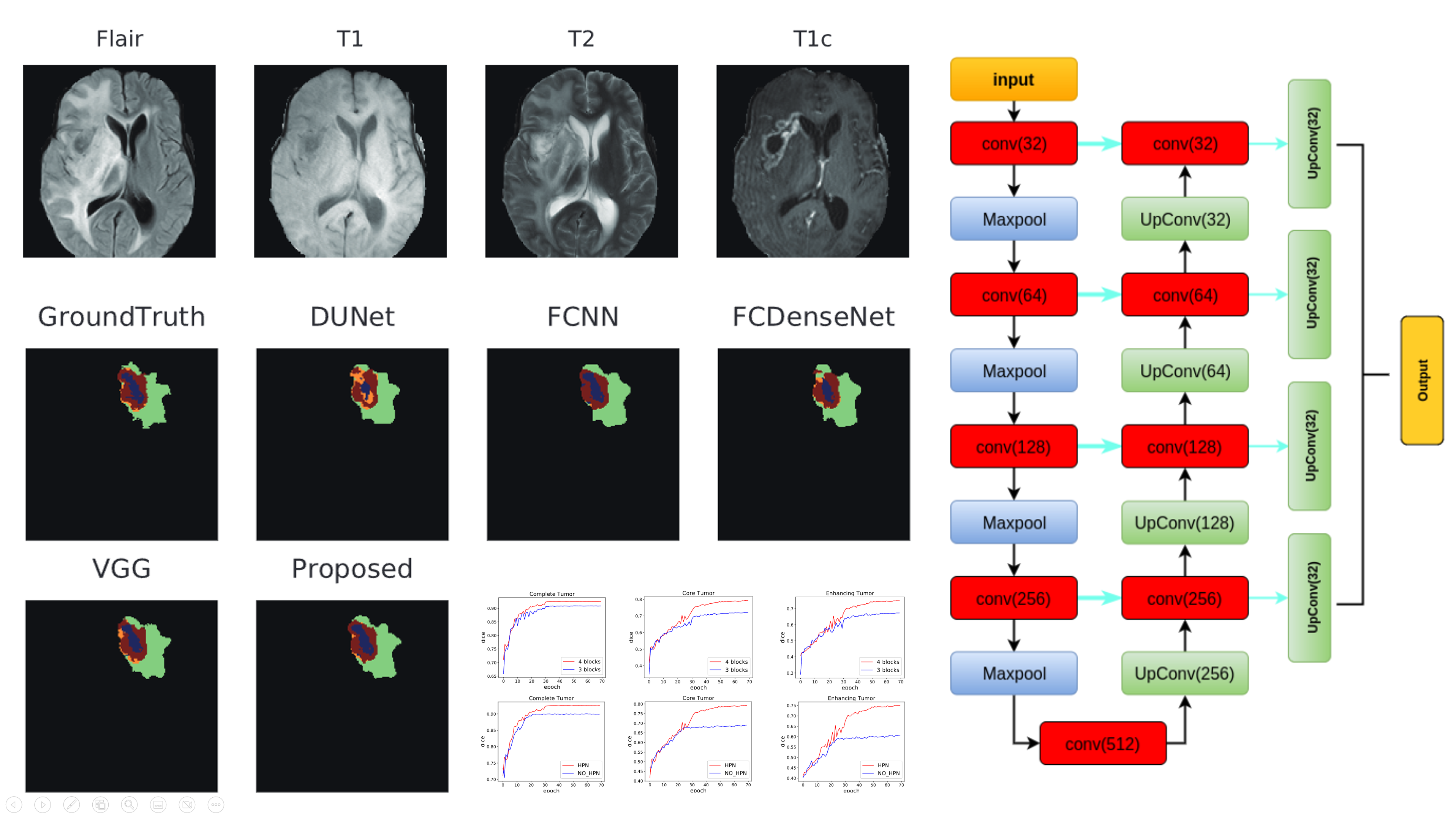 (2018). Hybrid pyramid u-net model for brain tumor segmentation. International conference on intelligent information processing:346–355.
(2018). Hybrid pyramid u-net model for brain tumor segmentation. International conference on intelligent information processing:346–355. -
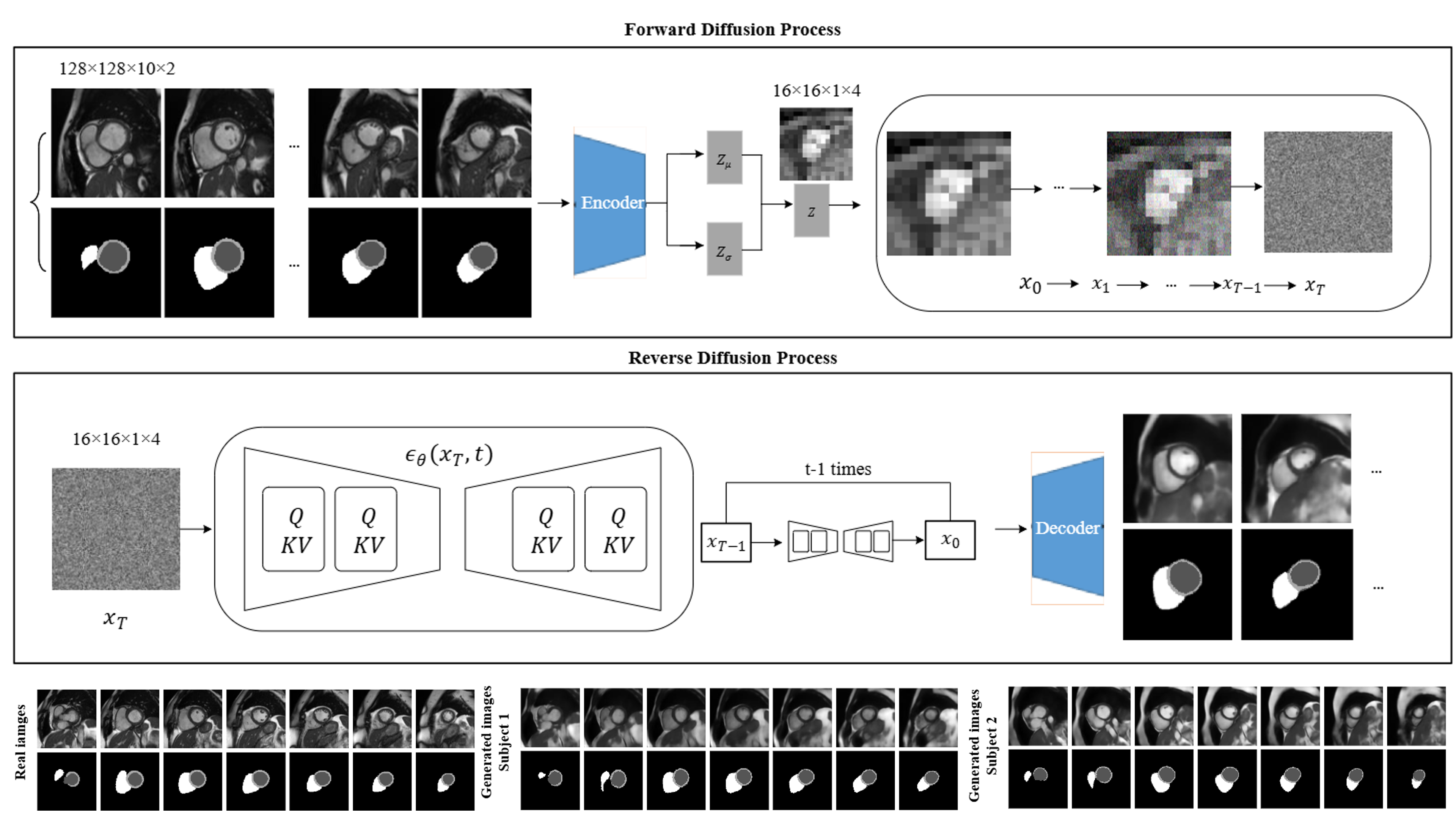 (2024). Synthesising 3D cardiac CINE-MR images and corresponding segmentation masks using a latent diffusion model. 2024 IEEE International Symposium on Biomedical Imaging (ISBI).
(2024). Synthesising 3D cardiac CINE-MR images and corresponding segmentation masks using a latent diffusion model. 2024 IEEE International Symposium on Biomedical Imaging (ISBI).